Research
Abey Research Group
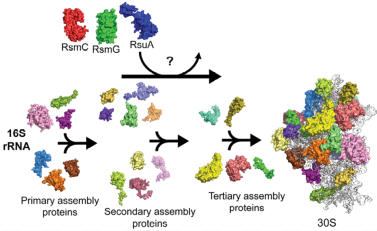
The
synthesis
of
new
ribosomes
is
critical
for
the
survival
of
all
organisms.
In
the
early
1960s,
Nomura
and
colleagues
found
30S
ribosomal
subunits
can
be
assembled
in
vitro,
and
the
assembly
is
highly
cooperative
and
hierarchical.
Since
then,
thermodynamics
and
kinetics
of
protein
addition,
RNA
folding
during
ribosomal
assembly,
and
the
structural
basis
for
cooperative
protein
addition
have
been
studied
extensively.
In
addition
to
r-proteins,
various
other
assembly
factors
and
nucleotide
modification
enzymes
are
involved
in
the
ribosomal
assembly.
Interestingly,
in
vivo,
both
ribosomal
assembly
and
rRNA
modifications
occur
co-transcriptionally.
However,
less
is
known
about
these
nucleotide
modification
steps
and
how
these
steps
are
integrated
into
the
mechanism
of
ribosomal
protein
addition.
Nucleotide
modification
steps
of
rRNA
can
influence
the
ribosomal
assembly
in
two
separate
ways.
First,
post-
transcriptional
modifications
can
perturb
local
RNA
structure,
stability,
and
dynamics
hence
modulating
RNA
folding.
Similarly,
modified
nucleotides
provide
additional
hydrophobic
patches
that
will
change
the
affinity
of
ribosomal
proteins.
Secondly,
thermodynamic
cooperativity
between
modification
enzymes
and
r-proteins
generates
low-energy
pathways
for
assembly
to
proceed.
Conversely,
kinetic
cooperativity
between
modification
enzymes
and
r-proteins
can
prevent
the
formation
of
undesirable
and
unproductive
assembly
intermediates.
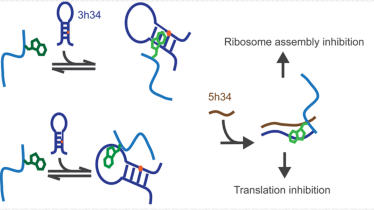
In
our
lab,
we
investigate
thermodynamic
and
kinetic
cooperativity
of
binding
between
rRNA
modification
enzyme and r-proteins.
read more...
Investigation of the ability of rRNA modification enzymes to influence
30S ribosome assembly
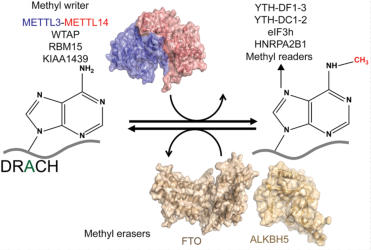
Nucleotide
modifications
such
as
N6-methyladenosines
(m
6
A),
5-
methylcytosines,
and
pseudouridines
are
observed
throughout
the
eukaryotic
transcriptome.
The
levels
of
nucleotide
modifications
vary
under
various
stress
conditions,
including
heat
shock,
starvation,
and
oxidative
stress.
These
observed
changes
in
mRNA
modification
levels
have
shown
to
control
viral
infections,
sperm
maturation,
and
cancer
progression.
In
cells,
m
6
A
levels
are
controlled
by
methyl
writers
and
erasers.
Methyl
reader
proteins
recognize
methylations
and
facilitate
mRNA
editing,
processing,
degradation,
or
increase
translation.
In
the
Abey
lab,
we
study
how
methyl
readers
recognize
methylated mRNA using various biochemical and biophysical techniques.
read more...

Study of N6-methyladenosine recognition by methyl readers
Since
the
discovery
of
penicillin,
many
antibiotic-resistant
bacterial
strains
have
emerged
all
throughout
the
world.
According
to
the
World
Health
Organization
(WHO),
antibiotic
resistance
of
bacteria
has
become
a
global
health
crisis.
Multidrug-resistant
Mycobacterium
tuberculosis
has
led
to
TB
infections, which alone have caused more than 250,000 deaths in 2015.
Ribosomes
are
the
target
for
many
classes
of
antibiotics.
These
diverse
classes
of
antibiotics
target
the
initiation,
elongation,
and
termination
steps
of
translation
in
bacteria.
The
correlation
between
nucleotide
modification
enzymes,
ribosome
assembly,
and
antibiotic
resistance
is
well
known.
For
example,
the
absence
of
methyltransferase
enzyme
KsgA,
cause
both
ribosome
assembly
defects
and
resistance
towards
kasugamycin.
In
the
Abey
lab,
we
investigate
the
potential
of
nucleotide
modification
machinery
as
a
potential drug target.
read more...
Discovery of small-molecular inhibitors of modification enzymes